Abstract
Background
Metabolic reprogramming is a major hallmark of cancer, providing tumor cells with vital energy and metabolites. Diffuse large B-cell lymphoma (DLBCL) is metabolically heterogeneous, and there was also evidence that altered metabolism is associated with tumor progression and prognosis. This study was aimed to assess the characteristics of metabolic reprogramming and novel biomarkers in DLBCL, thereby proposing a novel therapeutic strategy for DLBCL.
Methods
Serum sample were obtained from100 patients with newly diagnosed DLBCL and 70 age-gender matched healthy controls in the study. Non-targeted metabolomics studies were performed based on liquid mass spectrometry (LC-MS) technology. The normalized peak areas (normalized by IS) metabolites were introduced into SIMCA software for multivariate statistical analysis. Metabolomic pathway analysis of metabolic biomarkers was performed using MetbraAnalyst5.0.
Results
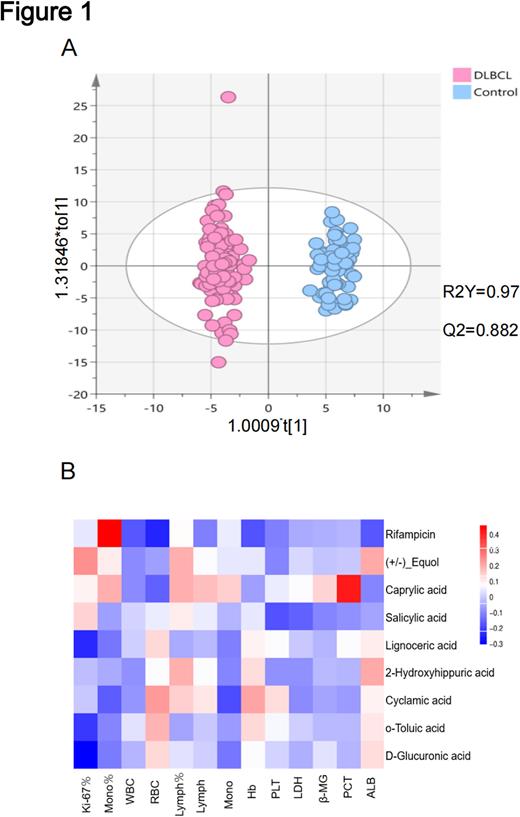
We first analyzed differential metabolite profiles in the serum of DLBCL patients. The metabolites detected in positive and negative ion modes were analyzed separately. Compared to the healthy control, DLBCL patients were featured with a specific metabolic profile, indicating the differential metabolism in disease state. We then established a model containing metabolic information by orthogonal partial least squares discriminant analysis (OPLS-DA) and tested its ability to identify new samples in seven cross-validations. The variable importance in the projection (VIP) was also calculated for each metabolite.
We further screened for differentially expressed metabolites by plotting collapsed changes (fold change>1.5 or <0.667, p<0.05). In DLBCL patients, 33 up-regulated metabolomes and 48 down-regulated metabolites were detected in positive ion mode, while 19 up-regulated metabolites and 29 down-regulated metabolites were in negative ion mode. Enrichment analyses showed that differential metabolites were mainly enriched in D-Glutamine and D-glutamate metabolism, caffeine metabolism, and ascorbate and aldarate metabolism.
We then attempted to verify whether the differential metabolites had diagnostic value in DLBCL patients. By using random forest analysis, 2-Phenylpropionicacid, 4-oxododecanedioicacid, cis-4-Hydroxy-D-proline, Hypoxanthine Octanedioicacid under positive ion mode, and Xanthosine, Subericacid, Diethyloxalpropionate, 4-Oxoproline Azelaicacid under negative ion mode, were screened out as potential biomarkers. Based on the above biomarkers, diagnostic models were developed and revealed the mean accuracies of 0.9460 in positive mode and 0.943 in negative mode. Receiver operating characteristic (ROC) curves then verified the significant sensitivity and specificity of the diagnostic models.
Notably, the Area Under Curve(AUC)of the combined diagnostic model was superior to that of each metabolite, indicating that combined model values could significantly improve the diagnostic performance in DLBCL.
Further investigations were aimed to explore the correlation of metabolite profiles with clinical features in DLBCL. Venn diagram profiled the differential metabolites between patients with GCB-like DLBCL and ABC-like DLBCL. Correlation analysis then suggested that the differential metabolites were correlated with clinical prognostic indicators in DLBCL patients. In particular, D-Glucuronic acid and Lignoceric acid were significantly associated with Ki-67 expression (tumor proliferation index), cis-4-Hydroxy-D-proline was significantly associated with LDH and β2-MG, and 2-Phenylpropionic acid significantly correlated with LDH, PLT and ALB.
Conclusions
Our study revealed the metabolic profile of DLBCL and found a relatively high diagnostic value of metabolite-based diagnostic models constructed. Furthermore, the combination with clinical data suggests that D-glucuronide, lignoceric acid, cis-4-hydroxy-D-proline and 2-phenylpropionic acid may be potential prognosis-related biomarkers in DLBCL. Targeting metabolic reprogramming is expected to provide novel strategies for the treatment of DLBCL.
Keywords
DLBCL, metabolomics, biomarkers, D-glucuronide, tumor
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.